Refactoring biology project : Part 2
Hi again!
This blog article is a follow up on my refactoring attempt of a biology project. Go read part 1 if you have missed it!
First task was to make the code more readable and easier to modify. Without changing how the variable were stored or the logic of the program, i just made helper function of the repetition. So, for the code of the earlier blog post:
def get_means(data):
temp_list = data
#Take out concentration values
concentrations = temp_list[:,0]
#Remove the blanks
concentrations = concentrations[np.logical_not(np.isnan(concentrations))]
nan_list=np.where(np.isnan(temp_list))
temp_list[nan_list]=np.take(concentrations, nan_list[1])
mean_value = np.mean(temp_list, axis=0)
x_value = temp_list[:,0]
y_value = temp_list[:,1]
return x_value, y_value, mean_value
To note: This is code i used during the refactor and not final version of it.
Ok it’s ugly, not so understandable but it does the job for now. Let’s just put it in a for loop and remove the old code. What behind the code doesn’t really matter in the scope of this blog post, but those functions are meant to read an excel file, associate duplicate or triplicates of a same measure to a concentration (x against y ) and save them for calculus and plotting later.
for elements in list_of_slices:
x, y , mean = helper.get_means(elements)
list_of_x.append(x)
list_of_y.append(y)
list_of_superior_y[index].append(y)
list_of_meanind[index].append(mean)
index += 1
ax1.plot(list_of_x, list_of_y, 'ko', markersize=5)
With just this modifications, and progressing bit by bit, i was able to reduce more than 500 lines of code and make it much more understandable. When this was done, i could have just stopped there but i decided that, if i wanted to make it a bit more clean and easy to maintain/expand, i should use object oriented programming.
For the abstraction of the problem, i considered that a triplicate of a reading, their means (the x if you plot it), and it’s associated concentration (the y) should be only one object. It makes accessing to a particular value easier. I tried to keep the logic and the handling of numpy arrays from the original author as close as possible to the initial design. (It would have been much easier to dump all files in dataframe, select the data we want and plot them easily).
I created so a new class called “Point”, because i’m very imaginative with names.
class Point():
"""
Each point is associated to a concentration,
coordinates (x, y), and mean
"""
def __init__(self, data):
self._concentration = data[:, 0][np.logical_not(np.isnan(data[:, 0]))]
data[np.where(np.isnan(data))] = np.take(self._concentration,
np.where(np.isnan(data))[1])
self._mean = np.mean(data, axis=0)
self._x = data[:, 0]
self._y = data[:, 1]
As you can see, it’s pretty much the same than the first function of this article and it act the same. But now, it is morehandy. Later, we merge all the points from different files. So for example, the point at 0.5 concentration from file A and B will get an average. So i created a Final_point class, because i’m very good with names.
But why stop the good works there? Each file, is a collection of Points coming from a csv file. Thoses files are separated in the file you wanna test and file of reference. So we can create an class that will parse the CSV files and be a collection of Point and Final_point.
class File_Parser:
"""Create an object from datas that is a list of Points objects from the
different files fed to it.
Attributes:
file_names (list): List of the files used to generate datas.
list_of_points (list): List of the different points across a file.
For more info, see the Point class.
list_of_final_point (list): List of "final points", for all the files in
file_names aggregate all the points corresponding to one given
concentration.
list_of_conc (list): List of each concentration used. For now, it wont
work if the files used to create data have different concentrations
across them.
"""
def __init__(self, input_dic, data=None, user_input=None):
"""Initialise a File_Parser object from a list of files
Args:
input_dic (dict): Dictionary of user inputs(given by GUI)
data (str): A string with the type of data. For now, only 'csv' is
supported. Pandas compatible file will be explored later.
user_input (str): A string for input_dict. Original plan was to
take all file in the same folder that finish with the same text. For
example 'Plate1good' and 'Plate2good" will be parsed together in a
File_Parser object and 'good' should be user_input.
As the program (for now) is built, we expect userinput1 or
userinput2.
"""
# Init lists
self.file_names = []
self.list_of_points = []
self.list_of_final_point = []
self.list_of_conc = []
if data == 'csv':
for files in glob.glob("*%s.csv" % user_input):
self.file_names.append(files)
# This is the delimitation used in the original repository and
# the files i'm using to test the program. We could extend the
# program to let user choose the data input (here it's array)
array = np.genfromtxt(files, delimiter=';',
skip_header=input_dic['user_input3']-1,
usecols=(input_dic['user_input4']-1,
input_dic['user_input5']-1))
list_of_slices = [array[0:3], array[3:6], array[6:9], array[9:12],
array[12:15], array[15:18], array[18:21],
array[21:24]]
for elements in list_of_slices:
point = Point(elements)
self.list_of_points.append(point)
if point.mean[0] not in self.list_of_conc:
self.list_of_conc.append(point.mean[0])
elif data == 'pandas':
# Pandas handling is planned. Pandas is much more easier to use
# than random arrays and would require an formated input with clear
# labels on values.
pass
else:
print('Unknown data format')
self.mask = list(range(len(self.file_names)))
# Take all points of a same concentration and average them into the
# Final_point class
for concentration in self.list_of_conc:
dummy_list = []
for point in self.list_of_points:
if point.mean[0] == concentration:
dummy_list.append(point)
self.list_of_final_point.append(Final_point(dummy_list))
And now the program is correct. It can also be more easily expanded and corrected upon.
And it seem to work :
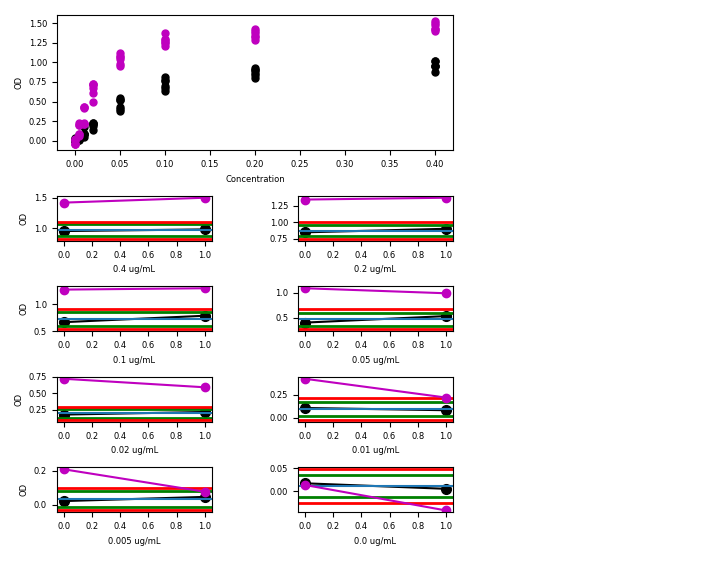
At the time i write this line, my pull request has not been accepted. You can find my full refactoring on github here. I could expand the project by making the data selection easier, pandas compatible or other things but i have also other projects as you will see in future blog posts ; )